Hands-on with socio4health: socioeconomic and demographic variables on dengue incidence in Colombia#
Run the tutorial via free cloud platforms:
This notebook provides you with a real world example on how to use socio4health to retrieve, harmonize and analyze socioeconomic and demographic variables related to dengue incidence in Colombia and recreate the dataset used in the publication Exploring Dengue Dynamics: A Multi-Scale Analysis of Spatio-Temporal Trends in Ibagué, Colombia by Otelo et al., published in Virus in 2024 (DOI). This tutorial assumes you have an intermediate or advanced understanding of Python and data manipulation.
Setting up the environment#
To run this notebook, you need to have the following prerequisites:
Python 3.10+
Additionally, you need to install the socio4health and pandas package, which can be done using pip:
!pip install socio4health pandas -q
[notice] A new release of pip is available: 25.1.1 -> 25.2
[notice] To update, run: python.exe -m pip install --upgrade pip
In case you want to run this notebook in Google Colab, you also need to run the following command to use your files stored in Google Drive:
from google.colab import drive
drive.mount('/content/drive')
Import Libraries#
To perform the data extraction, the socio4health library provides the Extractor class for data extraction, and the Harmonizer class for data harmonization of the retrieved date. pandas will be used for data manipulation. Additionally, we will use some utility functions from the socio4health.utils.harmonizer_utils module to standardize and translate the dictionary.
import datetime
import geopandas as gpd
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from socio4health import Extractor
from socio4health.harmonizer import Harmonizer
from socio4health.utils import harmonizer_utils
1. Extract data from Colombia#
To extract data from Colombia, use the Extractor class from the socio4health library. As in the publication, extract the Colombian National Population and Housing Census 2018 (CNPV 2018) dataset from the Colombian Nacional Administration of Statistics (DANE) website. The dataset is available at: https://microdatos.dane.gov.co/index.php/catalog/643/related-materials.
The Extractor class requires the following parameters:
input_path: TheURLor local path to the data source.down_ext: A list of file extensions to download. This can include.CSV,.csv,.zip, etc.output_path: The local path where the extracted data will be saved.key_words: A list of keywords to filter the files to be downloaded. In this case, we are only interested in the file14045.zip, which contains the data at the desired level of granularity (census block level or “Manzana”).depth: The depth of the directory structure to traverse when downloading files. A depth of0means only the files in the specified directory will be downloaded.
col_online_extractor = Extractor(input_path="https://microdatos.dane.gov.co/index.php/catalog/643/related-materials",
down_ext=['.cpg', '.dbf', '.prj','.sbn', '.sbx', '.shx', '.shp', '.zip'],
output_path="../CNVP2018",
key_words=["14045.zip"],
depth=0)
col_CNPV = col_online_extractor.s4h_extract()
2025-09-23 09:49:52,233 - INFO - ----------------------
2025-09-23 09:49:52,235 - INFO - Starting data extraction...
2025-09-23 09:49:52,236 - INFO - Extracting data in online mode...
2025-09-23 09:49:52,236 - INFO - Scraping URL: https://microdatos.dane.gov.co/index.php/catalog/643/related-materials with depth 0
2025-09-23 09:49:56,948 - INFO - Spider completed successfully for URL: https://microdatos.dane.gov.co/index.php/catalog/643/related-materials
2025-09-23 09:49:56,950 - INFO - Downloading files to: ../CNVP2018
Downloading files: 0%| | 0/1 [00:00<?, ?it/s]2025-09-23 09:50:07,974 - INFO - Successfully downloaded: 14045.zip
Downloading files: 100%|██████████| 1/1 [00:11<00:00, 11.02s/it]
2025-09-23 09:50:07,986 - INFO - Processing (depth 0): 14045.zip
2025-09-23 09:50:15,101 - INFO - Extracted: 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_ANM_MANZANA.cpg
2025-09-23 09:50:17,199 - INFO - Extracted: 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_ANM_MANZANA.dbf
2025-09-23 09:50:17,204 - INFO - Extracted: 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_ANM_MANZANA.prj
2025-09-23 09:50:17,219 - INFO - Extracted: 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_ANM_MANZANA.sbn
2025-09-23 09:50:17,227 - INFO - Extracted: 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_ANM_MANZANA.sbx
2025-09-23 09:50:17,862 - INFO - Extracted: 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_ANM_MANZANA.shp
2025-09-23 09:50:17,886 - INFO - Extracted: 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_ANM_MANZANA.shx
Processing files: 0%| | 0/7 [00:00<?, ?it/s]2025-09-23 09:50:17,904 - WARNING - Unsupported extension: .cpg
2025-09-23 09:50:17,906 - WARNING - Unsupported extension: .shx
2025-09-23 09:50:17,909 - WARNING - Unsupported extension: .dbf
2025-09-23 09:50:17,917 - WARNING - Unsupported extension: .prj
Processing files: 71%|███████▏ | 5/7 [01:13<00:29, 14.60s/it]2025-09-23 09:51:31,124 - WARNING - Unsupported extension: .sbn
Processing files: 86%|████████▌ | 6/7 [01:13<00:11, 11.41s/it]2025-09-23 09:51:31,132 - WARNING - Unsupported extension: .sbx
Processing files: 100%|██████████| 7/7 [01:13<00:00, 10.46s/it]
2025-09-23 09:51:31,136 - INFO - Successfully processed 7/7 files
2025-09-23 09:51:31,141 - INFO - Extraction completed successfully.
col_CNPV[0]
| COD_DANE_A | DPTO_CCDGO | MPIO_CCDGO | MPIO_CDPMP | CLAS_CCDGO | SETR_CCDGO | SETR_CCNCT | SECR_CCDGO | SECR_CCNCT | ZU_CCDGO | ... | TP51_13_ED | TP51_99_ED | CD_LC_CM | NMB_LC_CM | TP_LC_CM | Shape_Leng | Shape_Area | COD_RDTM | geometry | filename | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0500210000000000010101 | 05 | 002 | 05002 | 1 | 000 | 050021000 | 00 | 05002100000 | 000 | ... | 10.0 | 1.0 | None | None | None | 0.002298 | 2.038760e-07 | 050021990000000000010101 | POLYGON ((-75.42779 5.79423, -75.4278 5.79422,... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
| 1 | 0500210000000000010102 | 05 | 002 | 05002 | 1 | 000 | 050021000 | 00 | 05002100000 | 000 | ... | 19.0 | 4.0 | None | None | None | 0.003402 | 5.600867e-07 | 050021990000000000010102 | POLYGON ((-75.42719 5.79421, -75.42715 5.79415... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
| 2 | 0500210000000000010103 | 05 | 002 | 05002 | 1 | 000 | 050021000 | 00 | 05002100000 | 000 | ... | 6.0 | 1.0 | None | None | None | 0.002622 | 4.293780e-07 | 050021990000000000010103 | POLYGON ((-75.42804 5.79294, -75.42807 5.79291... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
| 3 | 0500210000000000010104 | 05 | 002 | 05002 | 1 | 000 | 050021000 | 00 | 05002100000 | 000 | ... | 11.0 | 2.0 | None | None | None | 0.002673 | 4.493171e-07 | 050021990000000000010104 | POLYGON ((-75.42853 5.79348, -75.4286 5.79342,... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
| 4 | 0500210000000000010105 | 05 | 002 | 05002 | 1 | 000 | 050021000 | 00 | 05002100000 | 000 | ... | 0.0 | 0.0 | None | None | None | 0.001338 | 8.776894e-08 | 050021990000000000010105 | POLYGON ((-75.4291 5.79393, -75.4291 5.79393, ... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 504991 | 9977320030102700010102 | 99 | 773 | 99773 | 2 | 003 | 997732003 | 01 | 99773200301 | 027 | ... | 0.0 | 0.0 | None | None | None | 0.002270 | 2.451366e-07 | 997732990030102700010102 | POLYGON ((-69.85155 4.33427, -69.85149 4.33427... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
| 504992 | 9977320030102700010103 | 99 | 773 | 99773 | 2 | 003 | 997732003 | 01 | 99773200301 | 027 | ... | 0.0 | 0.0 | None | None | None | 0.002919 | 5.196303e-07 | 997732990030102700010103 | POLYGON ((-69.85227 4.33365, -69.85257 4.3337,... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
| 504993 | 9977320030102700010104 | 99 | 773 | 99773 | 2 | 003 | 997732003 | 01 | 99773200301 | 027 | ... | 0.0 | 0.0 | None | None | None | 0.002938 | 3.280837e-07 | 997732990030102700010104 | POLYGON ((-69.85276 4.33338, -69.85274 4.3336,... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
| 504994 | 9977320030102700010105 | 99 | 773 | 99773 | 2 | 003 | 997732003 | 01 | 99773200301 | 027 | ... | 0.0 | 0.0 | None | None | None | 0.002514 | 3.375903e-07 | 997732990030102700010105 | POLYGON ((-69.85313 4.33348, -69.85311 4.33368... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
| 504995 | 9977320030102700010106 | 99 | 773 | 99773 | 2 | 003 | 997732003 | 01 | 99773200301 | 027 | ... | 0.0 | 0.0 | None | None | None | 0.002535 | 1.992453e-07 | 997732990030102700010106 | POLYGON ((-69.85425 4.33397, -69.8541 4.33396,... | 383d6920_MGN_NivelManzana_Integrado_CNPV_MGN_A... |
504996 rows × 111 columns
2. Load and standardize the dictionary#
To harmonize the data, provide a dictionary that describes the variables in the dataset. The CNPV 2018 dictionary is available at here. Follow the steps in the tutorial “How to Create a Raw Dictionary for Data Harmonization” to create a raw dictionary in Excel format.
raw_dic = pd.read_excel("raw_dic_mzn.xlsx")
raw_dic
| variable_name | type | size | question | description | value | |
|---|---|---|---|---|---|---|
| 0 | COD_DANE_A | Text | 22.0 | Código de manzana concatenado (departamento, m... | NaN | NaN |
| 1 | DPTO_CCDGO | Text | 2.0 | Código del departamento | NaN | NaN |
| 2 | MPIO_CCDGO | Text | 3.0 | Código del municipio | NaN | NaN |
| 3 | MPIO_CDPMP | Text | 5.0 | Código concatenado que identifica al municipio | NaN | NaN |
| 4 | CLAS_CCDGO | Text | 1.0 | Código de la clase 1 cabecera municipal, 2 cen... | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... |
| 101 | TP51_13_ED | Double | NaN | Conteo de personas donde el nivel educativo de... | Ninguno | NaN |
| 102 | TP51_99_ED | Double | NaN | Conteo de personas donde el nivel educativo de... | Sin información | NaN |
| 103 | CD_LC_CM | Text | 10.0 | Código de la localidad o comuna | NaN | NaN |
| 104 | NMB_LC_CM | Text | 50.0 | Nombre de la localidad o comuna | NaN | NaN |
| 105 | TP_LC_CM | Text | 20.0 | Descripción de tipo localidad o comuna o corre... | NaN | NaN |
106 rows × 6 columns
Standardize the dictionary using the s4h_standardize_dict function from the harmonizer_utils module. Then, translate it to English using the s4h_translate_column function from the same module (Note: This function depends on your internet connection and Google’s deep_translator extension, so it may take a few minutes). Finally, classify the variables into categories using a pre-trained BERT model.
dic = harmonizer_utils.s4h_standardize_dict(raw_dic)
dic = harmonizer_utils.s4h_translate_column(dic, "question", language="en")
dic = harmonizer_utils.s4h_translate_column(dic, "description", language="en")
dic = harmonizer_utils.s4h_translate_column(dic, "possible_answers", language="en")
C:\Users\isabe\PycharmProjects\socio4health\src\socio4health\utils\harmonizer_utils.py:98: FutureWarning: DataFrameGroupBy.apply operated on the grouping columns. This behavior is deprecated, and in a future version of pandas the grouping columns will be excluded from the operation. Either pass `include_groups=False` to exclude the groupings or explicitly select the grouping columns after groupby to silence this warning.
.apply(_process_group, include_groups=True)\
question translated
description translated
possible_answers translated
dic = harmonizer_utils.s4h_classify_rows(dic, "question_en", "description_en", "possible_answers_en",
new_column_name="category",
MODEL_PATH="files/bert_finetuned_classifier")
Device set to use cpu
This is how the standardized and translated dictionary looks:
dic
| variable_name | type | size | question | description | value | possible_answers | question_en | description_en | possible_answers_en | category | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | VERSION | Long Integer | NaN | año de la información geográfica | NaN | NaN | NaN | year of geographical information | NaN | NaN | Identification |
| 1 | CTNENCUEST | Double | NaN | cantidad de encuestas cnpv 2018 | NaN | NaN | NaN | Number of CNPV 2018 surveys | NaN | NaN | Identification |
| 2 | TP3_1_SI | Double | NaN | cantidad de encuestas que reportaron estar en ... | NaN | NaN | NaN | number of surveys that reported to be in ethni... | NaN | NaN | Identification |
| 3 | TP3A_RI | Double | NaN | cantidad de encuestas que reportaron estar en ... | resguardo indígena | NaN | NaN | number of surveys that reported to be in ethni... | Indigenous shelter | NaN | Identification |
| 4 | TP3B_TCN | Double | NaN | cantidad de encuestas que reportaron estar en ... | tccn | NaN | NaN | number of surveys that reported to be in ethni... | TCCN | NaN | Identification |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 101 | DATO_ANM | Text | 50.0 | nombre capa anonimizada | NaN | NaN | NaN | Anonymity layer name | NaN | NaN | Identification |
| 102 | NMB_LC_CM | Text | 50.0 | nombre de la localidad o comuna | NaN | NaN | NaN | locality or commune name | NaN | NaN | Identification |
| 103 | TP27_PERSO | Double | NaN | número de personas | NaN | NaN | NaN | number of people | NaN | NaN | Identification |
| 104 | DENSIDAD | Double | NaN | número promedio de habitantes en la manzana qu... | NaN | NaN | NaN | Average number of inhabitants in the apple liv... | NaN | NaN | Identification |
| 105 | AREA | Double | NaN | área de la manzana en metros cuadrados (sistem... | NaN | NaN | NaN | Apple area in square meters (Magna_Colombia_bo... | NaN | NaN | Identification |
106 rows × 11 columns
3. Harmonize the data#
Use the Harmonizer class from the socio4health library to harmonize the data. First, set the similarity threshold to 0.9, meaning that only variables with a similarity score of 0.9 or higher will be considered for harmonization. Next, set the nan_threshold to 1, meaning that only variables with no missing values will be considered for harmonization.
The available columns in the dataset can be checked using the s4h_get_available_columns method. This method takes a list of DataFrames as input and returns a list of column names that are present in the DataFrames.
har = Harmonizer()
har.similarity_threshold = 0.9
har.nan_threshold = 1
available_columns = har.s4h_get_available_columns(col_CNPV)
available_columns
['AG_CCDGO',
'AREA',
'CD_LC_CM',
'CLAS_CCDGO',
'COD_DANE_A',
'COD_RDTM',
'CTNENCUEST',
'DATO_ANM',
'DENSIDAD',
'DPTO_CCDGO',
'LATITUD',
'LONGITUD',
'MANZ_CCDGO',
'MPIO_CCDGO',
'MPIO_CDPMP',
'NMB_LC_CM',
'PERSONAS_L',
'PERSONAS_S',
'SECR_CCDGO',
'SECR_CCNCT',
'SECU_CCDGO',
'SECU_CCNCT',
'SETR_CCDGO',
'SETR_CCNCT',
'SETU_CCDGO',
'SETU_CCNCT',
'Shape_Area',
'Shape_Leng',
'TP14_1_TIP',
'TP14_2_TIP',
'TP14_3_TIP',
'TP14_4_TIP',
'TP14_5_TIP',
'TP14_6_TIP',
'TP15_1_OCU',
'TP15_2_OCU',
'TP15_3_OCU',
'TP15_4_OCU',
'TP16_HOG',
'TP19_ACU_1',
'TP19_ACU_2',
'TP19_ALC_1',
'TP19_ALC_2',
'TP19_EE_1',
'TP19_EE_2',
'TP19_EE_E1',
'TP19_EE_E2',
'TP19_EE_E3',
'TP19_EE_E4',
'TP19_EE_E5',
'TP19_EE_E6',
'TP19_EE_E9',
'TP19_GAS_1',
'TP19_GAS_2',
'TP19_GAS_9',
'TP19_INTE1',
'TP19_INTE2',
'TP19_INTE9',
'TP19_RECB1',
'TP19_RECB2',
'TP27_PERSO',
'TP32_1_SEX',
'TP32_2_SEX',
'TP34_1_EDA',
'TP34_2_EDA',
'TP34_3_EDA',
'TP34_4_EDA',
'TP34_5_EDA',
'TP34_6_EDA',
'TP34_7_EDA',
'TP34_8_EDA',
'TP34_9_EDA',
'TP3A_RI',
'TP3B_TCN',
'TP3_1_SI',
'TP3_2_NO',
'TP4_1_SI',
'TP4_2_NO',
'TP51POSTGR',
'TP51PRIMAR',
'TP51SECUND',
'TP51SUPERI',
'TP51_13_ED',
'TP51_99_ED',
'TP9_1_USO',
'TP9_2_1_MI',
'TP9_2_2_MI',
'TP9_2_3_MI',
'TP9_2_4_MI',
'TP9_2_9_MI',
'TP9_2_USO',
'TP9_3_10_N',
'TP9_3_1_NO',
'TP9_3_2_NO',
'TP9_3_3_NO',
'TP9_3_4_NO',
'TP9_3_5_NO',
'TP9_3_6_NO',
'TP9_3_7_NO',
'TP9_3_8_NO',
'TP9_3_99_N',
'TP9_3_9_NO',
'TP9_3_USO',
'TP9_4_USO',
'TP_LC_CM',
'TVIVIENDA',
'VERSION',
'ZU_CCDGO',
'ZU_CDIVI',
'filename',
'geometry']
Configure the dictionary, categories, additional columns, key column, and key values to prepare for the harmonization process. Set the dict_df parameter to the standardized and translated dictionary. Set the categories parameter to “Housing” to ensure that only housing-related variables are considered for harmonization. Set the extra_cols and key_colparameter to MPIO_CDPMP, which stands for the municipality code. This allows for filtering the data to include only records from a specific municipality. In this case, we are interested in the municipality with the code 73001, which corresponds to Ibague. Finally, use the s4h_data_selector method to filter the data based on the specified key column and key values. This method returns a list of DataFrames that match the filtering criteria.
col_CNPV[0].shape
(504996, 111)
har.dict_df = dic
har.categories = ["Housing"]
har.extra_cols = ['MPIO_CDPMP', 'GEOMETRY']
har.key_col = 'MPIO_CDPMP'
har.key_val = ['73001']
filtered_ddfs = har.s4h_data_selector(col_CNPV)
The method s4h_compare_with_dict can be used to compare the available columns in the dataset with the variables in the dictionary. This method helps to identify which variables from the dictionary are present in the dataset and can be harmonized.
Finally, display the filtered DataFrames to see the harmonized data for the specified municipality. If needed, the harmonized data can be exported to a CSV file using the to_csv method.
har.s4h_compare_with_dict(col_CNPV)
Matches with dict_df: 106 (95.50%)
| Unmatched ddfs variable | Unmatched dict_df variables | |
|---|---|---|
| 0 | COD_RDTM | None |
| 1 | FILENAME | None |
| 2 | GEOMETRY | None |
| 3 | SHAPE_AREA | None |
| 4 | SHAPE_LENG | None |
Finally, display the filtered DataFrames to see the harmonized data for the specified municipality. If needed, the harmonized data can be exported to a CSV file using the to_csv method or visualized using matplotlib, geopandas, and numpy as shown below:
filtered_ddfs
[ MPIO_CDPMP TP9_4_USO TP9_2_USO TP9_3_USO TP9_1_USO TP9_2_9_MI \
422086 73001 0.0 1.0 1.0 48.0 0.0
422087 73001 0.0 0.0 2.0 67.0 0.0
422088 73001 0.0 0.0 0.0 0.0 0.0
422089 73001 0.0 2.0 3.0 110.0 0.0
422090 73001 0.0 0.0 1.0 0.0 0.0
... ... ... ... ... ... ...
427978 73001 0.0 1.0 0.0 9.0 0.0
427979 73001 0.0 1.0 1.0 3.0 0.0
427980 73001 0.0 1.0 0.0 9.0 0.0
427981 73001 0.0 0.0 0.0 8.0 0.0
427982 73001 0.0 0.0 0.0 1.0 0.0
TP9_3_4_NO TP9_3_7_NO TP9_3_3_NO TP9_3_99_N ... TP19_INTE2 \
422086 0.0 0.0 0.0 0.0 ... 38.0
422087 0.0 0.0 0.0 0.0 ... 8.0
422088 0.0 0.0 0.0 0.0 ... 0.0
422089 0.0 0.0 1.0 0.0 ... 63.0
422090 0.0 0.0 0.0 0.0 ... 0.0
... ... ... ... ... ... ...
427978 0.0 0.0 0.0 0.0 ... 5.0
427979 0.0 0.0 0.0 0.0 ... 0.0
427980 0.0 0.0 0.0 0.0 ... 8.0
427981 0.0 0.0 0.0 0.0 ... 7.0
427982 0.0 0.0 0.0 0.0 ... 0.0
TP19_RECB2 TP14_2_TIP TP14_1_TIP TP14_6_TIP TP14_3_TIP \
422086 0.0 10.0 22.0 1.0 16.0
422087 1.0 57.0 10.0 0.0 0.0
422088 0.0 0.0 0.0 0.0 0.0
422089 3.0 81.0 20.0 0.0 11.0
422090 0.0 0.0 0.0 0.0 0.0
... ... ... ... ... ...
427978 4.0 1.0 9.0 0.0 0.0
427979 0.0 0.0 4.0 0.0 0.0
427980 4.0 0.0 10.0 0.0 0.0
427981 1.0 0.0 8.0 0.0 0.0
427982 0.0 0.0 1.0 0.0 0.0
TP14_4_TIP TP14_5_TIP TP15_3_OCU \
422086 0.0 0.0 1.0
422087 0.0 0.0 2.0
422088 0.0 0.0 0.0
422089 0.0 0.0 1.0
422090 0.0 0.0 0.0
... ... ... ...
427978 0.0 0.0 2.0
427979 0.0 0.0 1.0
427980 0.0 0.0 2.0
427981 0.0 0.0 1.0
427982 0.0 0.0 1.0
GEOMETRY
422086 POLYGON ((-75.25204 4.45339, -75.25203 4.45339...
422087 POLYGON ((-75.24798 4.44987, -75.24797 4.44985...
422088 POLYGON ((-75.24873 4.44883, -75.24877 4.44879...
422089 POLYGON ((-75.24897 4.44924, -75.24893 4.44919...
422090 POLYGON ((-75.25099 4.45195, -75.25111 4.45187...
... ...
427978 POLYGON ((-75.19101 4.35893, -75.191 4.35893, ...
427979 POLYGON ((-75.18923 4.35806, -75.18929 4.35799...
427980 POLYGON ((-75.19102 4.35539, -75.19116 4.35513...
427981 POLYGON ((-75.19008 4.35431, -75.18991 4.35423...
427982 POLYGON ((-75.19121 4.35258, -75.19168 4.35285...
[5897 rows x 42 columns]]
df = filtered_ddfs[0]
# Create a GeoDataFrame
gdf = gpd.GeoDataFrame(df, geometry='GEOMETRY', crs="EPSG:4326")
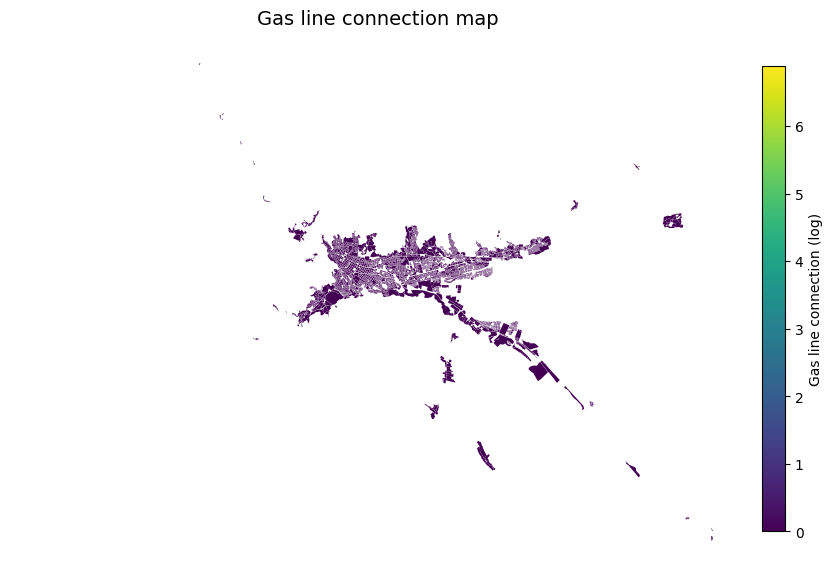
gdf['TP19_GAS_1_Log'] =np.log(gdf['TP19_GAS_1'])
C:\Users\isabe\PycharmProjects\socio4health\.venv\Lib\site-packages\pandas\core\arraylike.py:399: RuntimeWarning: divide by zero encountered in log
result = getattr(ufunc, method)(*inputs, **kwargs)
# Plot
fig, ax = plt.subplots(figsize=(10, 8))
# plot without automatic colorbar
gdf.plot(column='TP19_GAS_1_Log', cmap='viridis', ax=ax)
# Plot with custom colorbar
sm = plt.cm.ScalarMappable(cmap='viridis')
sm.set_array(gdf['TP19_GAS_1_Log'])
cbar = fig.colorbar(sm, ax=ax, fraction=0.03, pad=0.02) # fraction controls the size of the colorbar
cbar.set_label("Gas line connection (log)")
ax.set_title("Gas line connection map", fontsize=14)
ax.set_axis_off()
plt.show()
C:\Users\isabe\PycharmProjects\socio4health\.venv\Lib\site-packages\matplotlib\colors.py:2294: RuntimeWarning: invalid value encountered in subtract
resdat -= vmin
C:\Users\isabe\PycharmProjects\socio4health\.venv\Lib\site-packages\matplotlib\colors.py:2295: RuntimeWarning: invalid value encountered in divide
resdat /= (vmax - vmin)